ATOM-Seq® (Adapter Template Oligo-Mediated Sequencing) Technologies
A Revolutionary New DNA-Capture Method for Next Generation Sequencing
Technological Overview
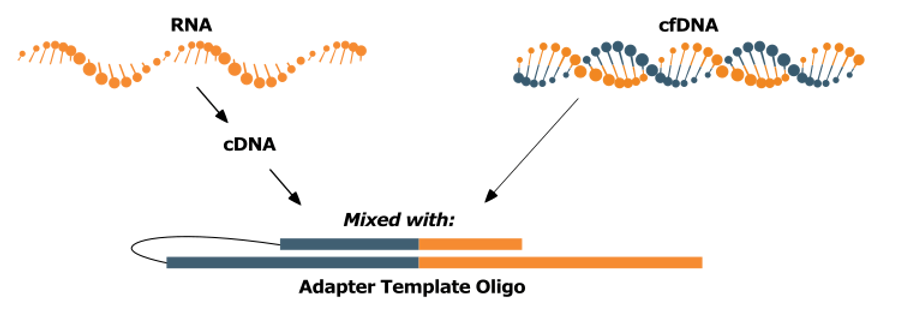
Our XCeloSeq NGS library preparation kits employ the novel, patented, GeneFirst breakthrough technology, ATOM-Seq, which uses a simple and elegant chemistry to capture DNA, where sample DNA molecules act as primers and their 3’ ends are extended by a polymerase.
ATOM-Seq incorporates Unique Molecular Identifiers (UMIs) and universal adapter sequences directly into available 3′ ends of every original sample molecule.
Step1. ATO Reaction:
Sample DNA or cDNA is combined with the Adapter Template Oligo
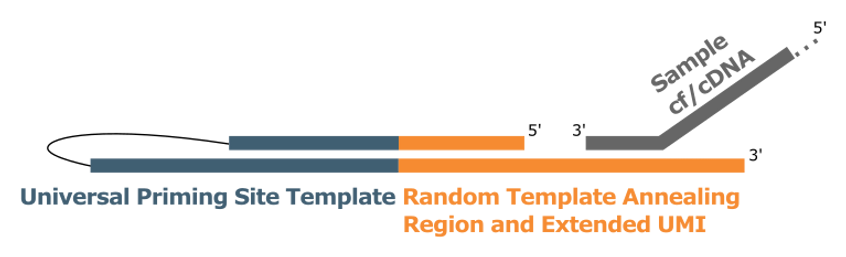
The DNA molecules anneal to the 3’ end of the ATO.
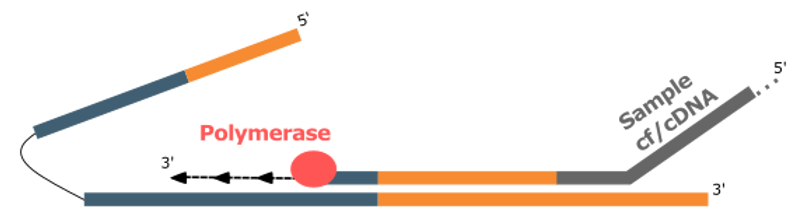
After annealing, a polymerase extends the original molecule, using the ATO sequence as a template.
Using its ATO template, the polymerase incorporates both a Unique Molecular Identifier (UMI) and a universal priming site into the captured molecule.
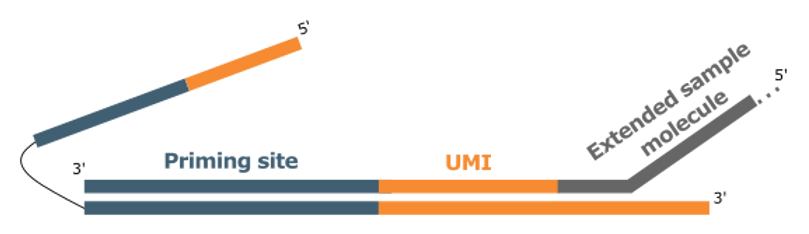
Step2. Linear Amplification
A universal primer is used for multiple rounds of linear amplification, improving error correction. This also means that all original copies can be represented in downstream reactions, even when the linear product is divided between sense- and antisense-strand targeted enrichment
| Downstream Protocols Protocol time includes all steps shown, from initial ATO Reaction to final, bead-purified library. | Targeted cfDNA Protocol Time: Total: 5h 40m Hands-on: 1h 20m Input Amount: Recommended: 5-50ng Minimum: 1ng Sensitivity: Allele frequency: ≥0.1% | Targeted RNA Protocol Time: Total: 7h 20m Hands-on: 1h 55m Input Amount: Recommended: 5-200ng Minimum: 1ng Both known and unknown gene fusions | Whole Genome Protocol Time: Total: 4h 45m Hands-on: 2h Input Amount: Recommended: 5-50ng Minimum: 1ng |
ATOM-Seq Key Advantages
This unique approach is ligation free and is set apart from conventional PCR-based approaches by requiring only a single target-specific primer for enrichment. This offers numerous advantages that make XCeloSeq particularly well suited to challenging clinical material such as cell-free DNA (cfDNA) from liquid biopsies or FFPE-preserved RNA.
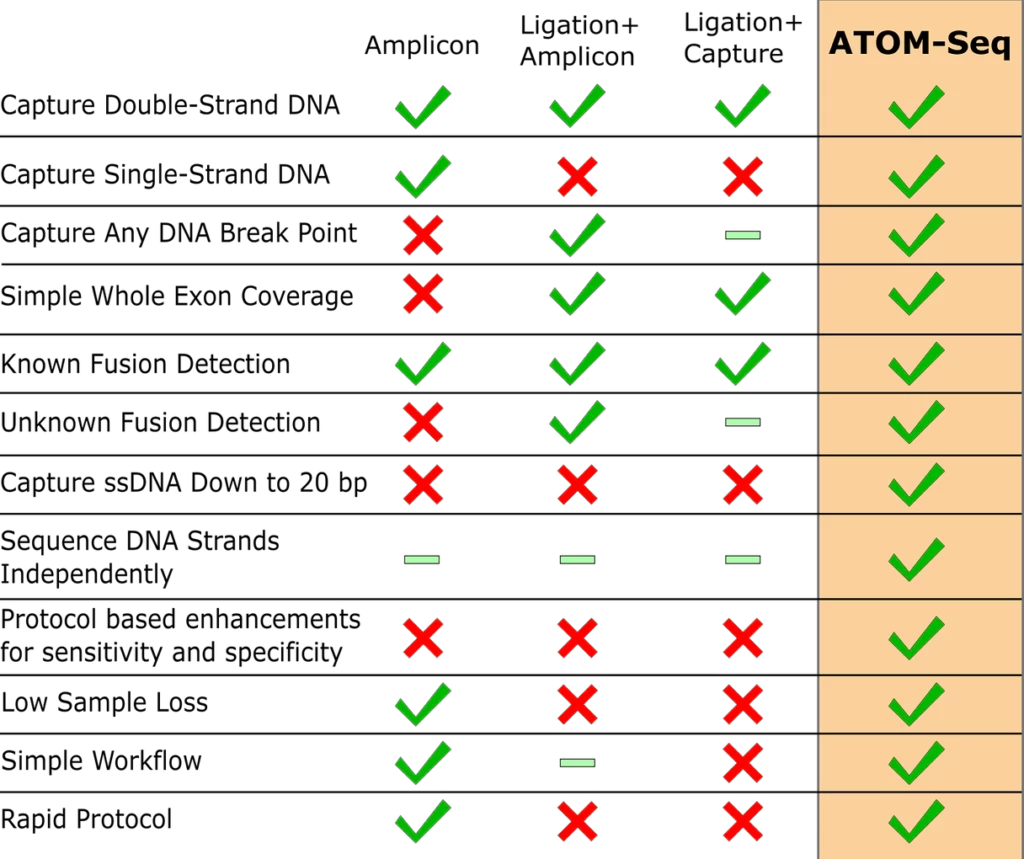
Green ticks indicate aspects the technologies can do.
Green bars indicate aspects of the technologies which are inefficient.
Red crosses indicate aspects the technologies cannot or have not been optimised to do.
Contact: huahong.wang@angenovo.no
For more info: https://www.genefirst.com/atom-seq-ngs
